156b Dnazyme-Based Nanosensors for Heavy Metals and Radionuclides
DNAzymes are biocatalysts with a promising capacity to selectively identify charged organic and inorganic compounds at ultratrace levels in industrial waste streams, chemical emissions, environmental samples or biological systems for a variety of applications. DNA enzymes are single-stranded DNA molecules with catalytic capabilities that are isolated from random-sequence DNA libraries by "in vitro selection". This new class of catalytic biomolecules has the potential of being used as unique molecular tools in a variety of innovative applications. Here, we describe the selection and characterization of an RNA-cleaving autocatalytic DNAzymes for heavy metals and radionuclides that links chemical catalysis with real-time fluorescence signaling in the single molecule. In vitro selection of RNA-cleaving DNAzymes which specifically use transition metal ions (Hg2+ and As5+) either through facilitation of the folding of DNA into stable tertiary structures, or through direct participation in the chemical reaction will be discussed over other ions and radionuclides.
We employed the specific multi-step selection scheme to isolate signaling autocatalytic DNA molecules. A pool of single-stranded 90-nt or 80-nt DNAs containing random-sequence nucleotides was first ligated to the acceptor DNA (23-nt) containing the three key moieties F (fluorophore; Fluorescein-dT), Ar (adenine ribonucleotide) and Q (quencher; DABCYL-dT). The ligated 123-nt or 113-nt DNA was purified by PAGE in step 2, followed by step 3 where the modified DNA molecules were incubated with several divalent metal ion cofactors for RNA cleavage. In the presence of metal ion, the enzyme carries out catalytic reaction of the substrate strand at the scissile ribonucleic acid adenosine (rA) resulting in removal of the quencher molecule and initiation of fluorescence. This will be measured using fluorescence spectroscopy. The selectivity of DNAzyme was monitored by change in fluorescence ((lex = 490 nm and lem = 520 nm). Any autocatalytic DNA capable of cleaving the lone RNA linkage was expected to generate either 102-nt or 92-nt DNA fragment that could be isolated by PAGE in step 4 and amplified over steps 5 and 6. The DNA product from the second PCR was treated with NaOH (step 7) under conditions that could fully cleave the ribonucleotide linkage (0.25 M NaOH, 90 0C, 10 min). The digested DNA mixture was subjected to PAGE purification and DNA phosphorylation (step 8). 5'-phosphorylated DNA was further used to initiate the next round of selection. Likewise, several cycles of selection and amplification were performed to isolate autocatalytic DNAzymes for specific metal ions by conducting the experiments in presence and absence of several metal ions. 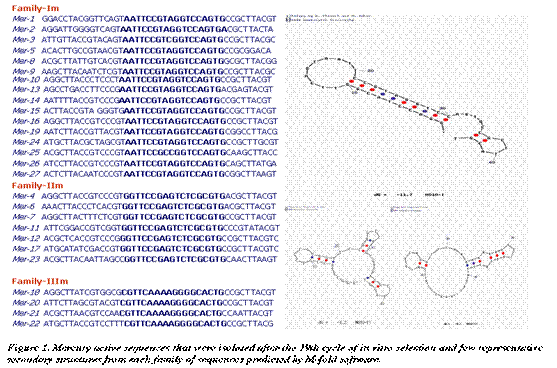 The reaction time and presence/absence of metal ion was then progressively evaluated to isolate very efficient DNAzymes for specific metal ions. The self-cleavage reaction (for Pool-A) was first allowed only to proceed for 10 min in C10 and 5 min in C11, and the reaction time was further reduced to 1 min in C12 and C13, to 10 s in C14 and C15, and finally to about 1 s in C16-C18. The DNA molecules in C19 were also allowed to react for 1 min. The activity of the selected DNA dramatically decreased from 10 min incubation (C10) to 1 min incubation (C12 & C13) leading to ca. 5 % and 7 % substrate cleavage respectively. Further gradual decrease in incubation time to 5 sec also produced minimal cleavage averaging ca. 1.5 % over three generations (C16-C18). At this stage, the incubation time was increased to 1 min (C19), resulting in a 50% cleavage efficiency. DNA sequences from the 19th cycle (C19) were amplified by PCR and cloned into a vector by the TA cloning method. The plasmids containing the individual catalysts were prepared using a Quiagen MiniPrep kit and sequenced.
The reaction time and presence/absence of metal ion was then progressively evaluated to isolate very efficient DNAzymes for specific metal ions. The self-cleavage reaction (for Pool-A) was first allowed only to proceed for 10 min in C10 and 5 min in C11, and the reaction time was further reduced to 1 min in C12 and C13, to 10 s in C14 and C15, and finally to about 1 s in C16-C18. The DNA molecules in C19 were also allowed to react for 1 min. The activity of the selected DNA dramatically decreased from 10 min incubation (C10) to 1 min incubation (C12 & C13) leading to ca. 5 % and 7 % substrate cleavage respectively. Further gradual decrease in incubation time to 5 sec also produced minimal cleavage averaging ca. 1.5 % over three generations (C16-C18). At this stage, the incubation time was increased to 1 min (C19), resulting in a 50% cleavage efficiency. DNA sequences from the 19th cycle (C19) were amplified by PCR and cloned into a vector by the TA cloning method. The plasmids containing the individual catalysts were prepared using a Quiagen MiniPrep kit and sequenced.
The population of molecules obtained after selection and reselection showed surprising similarity in their sequences when they were evolved using in vitro selection. The sequences of 27 individual clones revealed a diverse combination of functional motifs and are classified into three major families based on sequence similarities (Figure 1). Out of 27 mercury-active sequences, 16 fell in to one family (Family-Im), exhibiting ‘AATTCCGTAGGTCCAGTG' as a conserved region. The remaining 11 sequences could be classified in two different families (Family - IIm & IIIm) of seven and four sequences, respectively. A secondary structure for above mentioned sequence (most common structural motif) was predicted by the M-fold program (Zuker, 2003) and on the basis of this structure, we designed a trans-cleaving enzyme (called Mer-27) and examined further for its metal ion specificity and kinetics as mentioned below. Most of these sequences were found to be highly active in the presence of Hg2+ than in the presence of Mg2+, As5+, Cd2+ or Pb2+ when tested individually. The confirmation of its catalytic activity and the specificity was established in the following experiments.
The results obtained from Pool-B sequences were found to be different from what was noticed with Pool-A. No cleavage was observed until the 8th cycle (C8) and progressively increased to 18% at C11 without further improvement even after three more cycles (C12-C14). When the incubation time was reduced to 10 min (C15), 5 min (C16), 1 min (C17-C18), 10 sec (C19-C20) and finally 1 sec (C21-C22) at 22nd cycle - over 8 cycles - the decrease in ribosomal cleavage to 2% was noticed at the lowest incubation time (1 sec). When increased to 1 min in the 23rd cycle, an increase in ribosomal cleavage to 14% was observed similar to Pool-A. DNA sequences from the 23rd generation were amplified, cloned into a vector by the TA cloning method and sequenced as mentioned earlier.
The functional molecules for Pool-B obtained from M-fold program (after selection and reselection) showed minimal dissimilarity leading to two families based on sequence similarities (Figure 2). The sequences of 17 individual clones revealed a major conserved motive ‘ATCTCCTCCTGTTC' found in 12 clones (family-Ia). The remaining five clones fell in to one family (Family-IIa). Also, the most common structural motif (called Ars-17) was engineered in to a trans-cleaving enzyme as mentioned above, and examined further for its metal ion specificity and kinetics. The activities found in both pools were lost once the respective metal ions were omitted from the reaction mixture.  The isolation of two distinct families of DNAzymes that recognize Hg2+ and As5+ resulted in the emergence of new functional motifs with high catalytic rates of 2.6 min-1 for Mer-27 and 1.7 min-1 for Ars-17. Independent of whether DNAzymes exist in nature, it may now be possible to create a wide range of engineered DNA enzymes for application as highly stable biocatalysts. The information obtained in this study for metal-specific DNAzymes can be used more efficiently to understand sequence, structural and functional configurations of DNA folding and hence design of effective DNAzyme-based biosensors for a variety of environmentally-relevant metal ligands. Combining the specificity of nano-biological recognition probes and the sensitivity of laser-based optical detection, DNAzymes are capable of recruiting transition metal ions to broaden the scope and increase the efficiency of biosensing capabilities. The present work on heavy metal recognition and future work on radionuclides will be presented in the context of detecting and differentiating chemical constituents of complex systems to provide unambiguous identification and accurate quantification.
The isolation of two distinct families of DNAzymes that recognize Hg2+ and As5+ resulted in the emergence of new functional motifs with high catalytic rates of 2.6 min-1 for Mer-27 and 1.7 min-1 for Ars-17. Independent of whether DNAzymes exist in nature, it may now be possible to create a wide range of engineered DNA enzymes for application as highly stable biocatalysts. The information obtained in this study for metal-specific DNAzymes can be used more efficiently to understand sequence, structural and functional configurations of DNA folding and hence design of effective DNAzyme-based biosensors for a variety of environmentally-relevant metal ligands. Combining the specificity of nano-biological recognition probes and the sensitivity of laser-based optical detection, DNAzymes are capable of recruiting transition metal ions to broaden the scope and increase the efficiency of biosensing capabilities. The present work on heavy metal recognition and future work on radionuclides will be presented in the context of detecting and differentiating chemical constituents of complex systems to provide unambiguous identification and accurate quantification.