156c Identification of Toxic Chemicals in Activated Sludge System by Denaturing High-Performance Liquid Chromatography
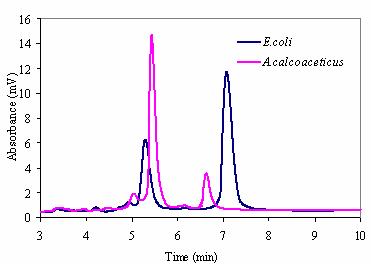
Introduction The activated sludge system is the most popular form of biological sewage treatment. However, shock loads of toxic chemicals represent a significant problem to activated sludge systems because they disrupt microbial metabolism resulting in a deterioration of process performance. Due to the significance of these disruptions on process performance, a number of research projects have been undertaken to develop biosensors (Microtox®, on-line respirometry) that are capable of identifying the source and the causative mechanism of an impeding upset. However, relatively few of these have progressed into commercial markets, even less have studied how the activated sludge microbial community responds to toxic chemicals. We HYPOTHESIZE that ribosome genesis can be used as a sensitive indicator of overall response to toxic chemicals. Ribosome genesis starts with the transcription of rrn operons to produce a polycistronic transcript, which is then processed in two steps by RNases to produce precursor rRNAs and then mature rRNAs. The rRNAs are finally combined with ribosomal proteins to produce a functional ribosome. It is well documented that high levels of pre16S rRNA correspond to the presence of a toxic inhibition in pure cultures as well as activated sludge. Recently, a novel reverse transcription and primer extension (RT&PE) approach was developed to investigate the secondary processing of precursor 16S rRNA for pure cultures by determining the ratio of pre16S-5' rRNA to 16S rRNA. This method was then tested successfully with activated sludge samples. The previous study suggested that the novel RT&PE method could be used to determine whether specific microbial populations are inhibited in activated sludge systems under shock loads of toxic chemicals. To expand this method, the current study examines the use of denaturing high performance liquid chromatography (DHPLC) by WAVE microbial analysis system (Transgenomic Ltd., Omaha, NE) to rapidly separate and identify signature in pre16S rRNA levels among mixed cultures of environmental bacteria. We used this approach to investigate the secondary processing of the 5' end of precursor 16S rRNA (pre16S-5' rRNA) for activated sludge exposed to copper (II) (Cu2+), 3, 5-dichlorophenol (3, 5-DCP), or 4-nitrophenol (4-NP). This study represents the first of its kind to document activated sludge response to toxic shock loadings using DHPLC-based quantification of pre16S rRNA levels. We aim to establish a routine approach to facilitate the identification of toxic materials in activated sludge system simply on the basis of the DHPLC profile obtained. In addition, it provides us with a better understanding of bacterial growth inhibition under toxic shock loading. Experimental Methods and Results Samples were collected from overnight, fresh growth media and fresh growth media with chloramphenicol which correspond to three distinct types of cells based on pre16S rRNA levels. RT & PE products from three cultures of A. calcoaceticus and Type III cells of A. calcoaceticus, E. coli, and M. mucogenicum shows that pre 16S ss DNA and 16S ss DNA were present as two prominent bands [400 and 550 nt]. This result conforms that under inhibition of growth, bacterial has higher levels of pre 16S rRNA as compared to the level of mature 16S rRNA. Although two distinct bands from the RT&PE product could be distinguished by agarose gel electrophoresis, the DHPLC-based approach using the WAVE system from Transgenomics shows promise for rapid throughput and sensitive quantification of single stranded DNA levels. RT&PE product that were derived from E. coli and A. calcoaceticus were analyzed on the WAVE system at 80.0°C (Figure 1). Both species exhibited distinct peak with characteristic retention times. The height of each particular peak was specified as the absorbance in mV and depended on the volume and the DNA concentration injected. This preliminary result with the WAVE system provides confidence that the approach described in this study can be used as a rapid and sensitive measure of microbial physiology. The current STATUS of this study is that we have successfully demonstrated that we can perform the RT&PE, and we have demonstrated that the WAVE system can be used to successfully quantify pre 16S rRNA levels. We have also generated samples of activated sludge collected from full-scale municipal treatment plants that have been exposed to toxic loads of copper and organic electrophiles. Currently, we are using the RT&PE approach coupled with the WAVE system to measure the physiological response of activated sludge to toxic loading. It is expected that the approach reported here can be used to establish a specialized database of bacterial phylogeny and to detect bacteria by the combination of distinct retention profiles of RT & PE product. We envision that this method will be used to understand how bacterial communities respond to toxic shock loadings.