152f Computer Aided Design of Modular Protein Devices: Logical "and" Gene Activation
When creating synthetic gene networks it is highly useful to turn on the expression of an engineered gene if and only if two different DNA-binding proteins exist in sufficient concentration. This type of behavior is referred to as AND logic and enables the creation of living biosensors and gene therapy systems which regulate production of a protein (either fluorescent or therapeutic) according to the presence of two separate protein inputs. The ideal biological AND gate consists of a gene or protein network that 1) only activates gene expression if both inputs are present, 2) has a low “false positive” rate of gene expression when one input is absent, 3) quickly responds to changes in the concentrations of the inputs, 4) contains a small number of components (as measured by DNA sequence length), and 5) contains only modular components so that, by choosing components from a library, one can create a library of AND gates that activate different genes with different regulatory protein inputs.
Using a quantitative model, we show how to build a protein device, or a system of interacting fusion proteins, that activates the transcriptional initiation of a specific gene if and only if two different DNA-binding proteins are present. The system contains DNA-binding, protein-protein interaction, non DNA-binding, and transactivating protein domains, fused together in a novel way. Importantly, each protein domain in the system lacks any cooperative or allosteric interactions and is completely replaceable by another of its type, drawn from a library of molecular components. Preliminary libraries of synthetic DNA-binding1 and protein-protein interaction2 domains have already been generated and natural versions of modular protein-protein interaction domains are also plentiful3. Consequently, the same design can be reused to create numerous distinct AND protein devices which may independently activate different genes with different regulatory inputs in the same cell.
The quantitative model combines a stochastic kinetic (Master equation) and thermodynamic description (partition function) of the protein-protein and protein-DNA interactions, respectively, using experimentally measured kinetic and thermodynamic data of existing protein domains. We use the model to predict the rate of transcriptional initiation as a function of the concentration of the two different DNA-binding proteins, including the false positive rate of transcriptional initiation. We then identify the molecular characteristics of each protein domain that yield a protein device with the most “pure” AND behavior and determine the response to dynamic changes in the inputs (see Figure for the dynamics of three different AND protein devices of varying purity).
Finally, we show how combining additional modular DNA-binding and protein-protein interaction domains can create protein devices that activate or repress transcriptional initiation with more complex logic, including compound AND.OR., OR.AND.OR, and AND.AND logical behaviors with 2, 3, or 4 regulatory inputs.
1 D. J. Segal et al., "Evaluation of a modular strategy for the construction of novel polydactyl zinc finger DNA-binding proteins," Biochemistry 42(7), 2137-48 (2003).
2 Astrid V. Giesecke, Rui Fang and J. Keith Joung, "Synthetic protein-protein interaction domains created by shuffling Cys2His2 zinc-fingers," Mol Syst Biol 2(1) (2006).
3 M. van Ham and W. Hendriks, "PDZ domains-glue and guide," Mol.Biol.Rep. 30(2), 69-82 (2003).
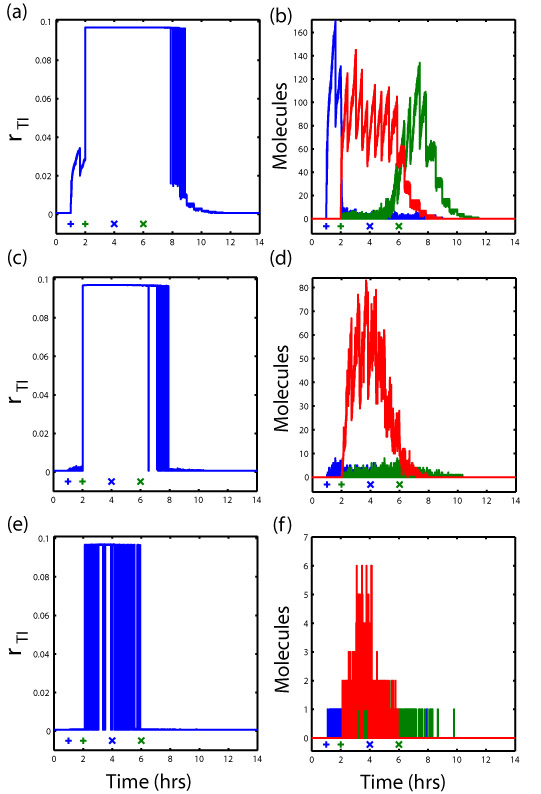
Figure Caption: (Left) The instantaneous rate of transcriptional initiation of a specific gene in response to dynamic changes in the production rates of the two input DNA-binding proteins. (+) Step up of production rate. (X) Step down of production rate to zero. (Right) The number of molecules of three forms of the protein device over time.